5.2. Useful Tips
Examples in this page assume that mp-22862 (NaCl) has been computed using auto-kappa.
Please also see auto-kappa/examples/6_parser/basic.
Make DFSET file for Alamode
#!/usr/bin/env python3
# Filename: make_dfset.py
import argparse
from auto_kappa.io.vasp import get_dfset
def main(options):
from plot_bandos import get_formula
formula = get_formula(options.base_dir)
dir_force = f"{options.base_dir}/cube/force_fd"
offset_xml = dir_force + '/prist/vasprun.xml'
outfile = 'DFSET.cube'
disps, force = get_dfset(dir_force, offset_xml=offset_xml, outfile=outfile)
if __name__ == '__main__':
parser = argparse.ArgumentParser(description='Input parameters')
parser.add_argument('--base_dir', dest='base_dir', type=str,
default="./mp-22862", help="directory of auto-kappa output")
args = parser.parse_args()
main(args)
# Filename: ./mp-22862/cube/force_fd/1/vasprun.xml, Snapshot: 1, E_pot (eV): -223.9017635
0.0566916869058 0.0000000000000 0.0000000000000 -2.4432377095235e-03 0.0000000000000e+00 -0.0000000000000e+00
0.0000000000000 0.0000000000000 0.0000000000000 5.8858366632325e-05 2.3052079147308e-04 -0.0000000000000e+00
0.0000000000000 0.0000000000000 0.0000000000000 5.8858366632325e-05 0.0000000000000e+00 2.3052079147308e-04
0.0000000000000 0.0000000000000 0.0000000000000 -6.4676878139020e-05 -7.5998472487173e-07 -7.5998472487173e-07
0.0000000000000 0.0000000000000 0.0000000000000 -1.9478011781769e-06 0.0000000000000e+00 0.0000000000000e+00
0.0000000000000 0.0000000000000 0.0000000000000 -1.3134434062906e-06 -3.0123243061062e-06 0.0000000000000e+00
0.0000000000000 0.0000000000000 0.0000000000000 5.8858366632325e-05 0.0000000000000e+00 -2.3052079147308e-04
0.0000000000000 0.0000000000000 0.0000000000000 -6.4676878139020e-05 -7.5998472487173e-07 7.5998472487173e-07
0.0000000000000 0.0000000000000 0.0000000000000 -1.9478011781769e-06 0.0000000000000e+00 -0.0000000000000e+00
Make BORNINFO file for ALAMODE
#!/usr/bin/env python3
# File: make_borninfo.py
import os
import argparse
import glob
from auto_kappa.io import BORNINFO
def main(options):
from plot_bandos import get_formula
formula = get_formula(options.base_dir)
file_vasprun = f"{options.base_dir}/nac/vasprun.xml"
file_fc2 = f"{options.base_dir}/harm/force/{formula}.xml"
# BORNINFO object creats BORNINFO file from vasprun.xml
# adjusting the order of atoms for ALAMODE force constants file.
born = BORNINFO(file_vasprun, file_fcs=file_fc2)
born.write(outfile="BORNINFO")
print("Primitive cell from vasprun.xml")
print("===============================")
print(born.prim_vasp)
print()
print("Primitive cell from ALAMODE force constants file")
print("================================================")
print(born.prim_fcs)
if __name__ == '__main__':
parser = argparse.ArgumentParser(description='Input parameters')
parser.add_argument('--base_dir', dest='base_dir', type=str,
default="./mp-22862", help="directory of auto-kappa output")
args = parser.parse_args()
main(args)
Plot phonon dispersion and DOS
#!/usr/bin/env python3
# Filename: plot_bandos.py
import os
import argparse
import glob
from auto_kappa.io import Band, Dos, BandPR
from auto_kappa.plot.initialize import set_matplot, set_axis
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
def get_formula(base_dir):
line = f"{base_dir}/harm/bandos/*.bands"
fns = glob.glob(line)
if len(fns) == 0:
raise ValueError(" No band file found")
fn = fns[0]
return os.path.basename(fn).split(".")[0]
def main(options):
fontsize = 7
fig_width = 3.5
aspect = 0.5
dpi = 600
## Prepare figure frame
set_matplot(fontsize=fontsize)
fig = plt.figure(figsize=(fig_width, aspect*fig_width))
plt.subplots_adjust(wspace=0.05)
gs = gridspec.GridSpec(1, 2, width_ratios=[3,1])
ax1 = fig.add_subplot(gs[0])
ax2 = fig.add_subplot(gs[1])
## Get file names
formula = get_formula(options.base_dir)
# file_band = f"{options.base_dir}/harm/bandos/{formula}.bands"
file_band_pr = f"{options.base_dir}/harm/bandos/{formula}.band.pr"
file_dos = f"{options.base_dir}/harm/bandos/{formula}.dos"
figname = "fig_bandos.png"
## Make Band and Dos objects
# band = Band(file_band)
band_pr = BandPR(file_band_pr)
dos = Dos(file_dos)
## Plot band and DOS
# band.plot(ax1, plot_G2G=False)
band_pr.plot(ax1, plot_G2G=False, cbar_location='upper left')
dos.plot(ax2, xlabel=None)
## Use same y limit
ax2.set_ylim(ax1.get_ylim())
## Adjust figure
ax2.tick_params(labelleft=False)
set_axis(ax1)
set_axis(ax2)
fig.savefig(figname, dpi=dpi, bbox_inches='tight')
print(" Output", figname)
if __name__ == '__main__':
parser = argparse.ArgumentParser(description='Input parameters')
parser.add_argument('--base_dir', dest='base_dir', type=str,
default="./mp-22862", help="directory of auto-kappa output")
args = parser.parse_args()
main(args)
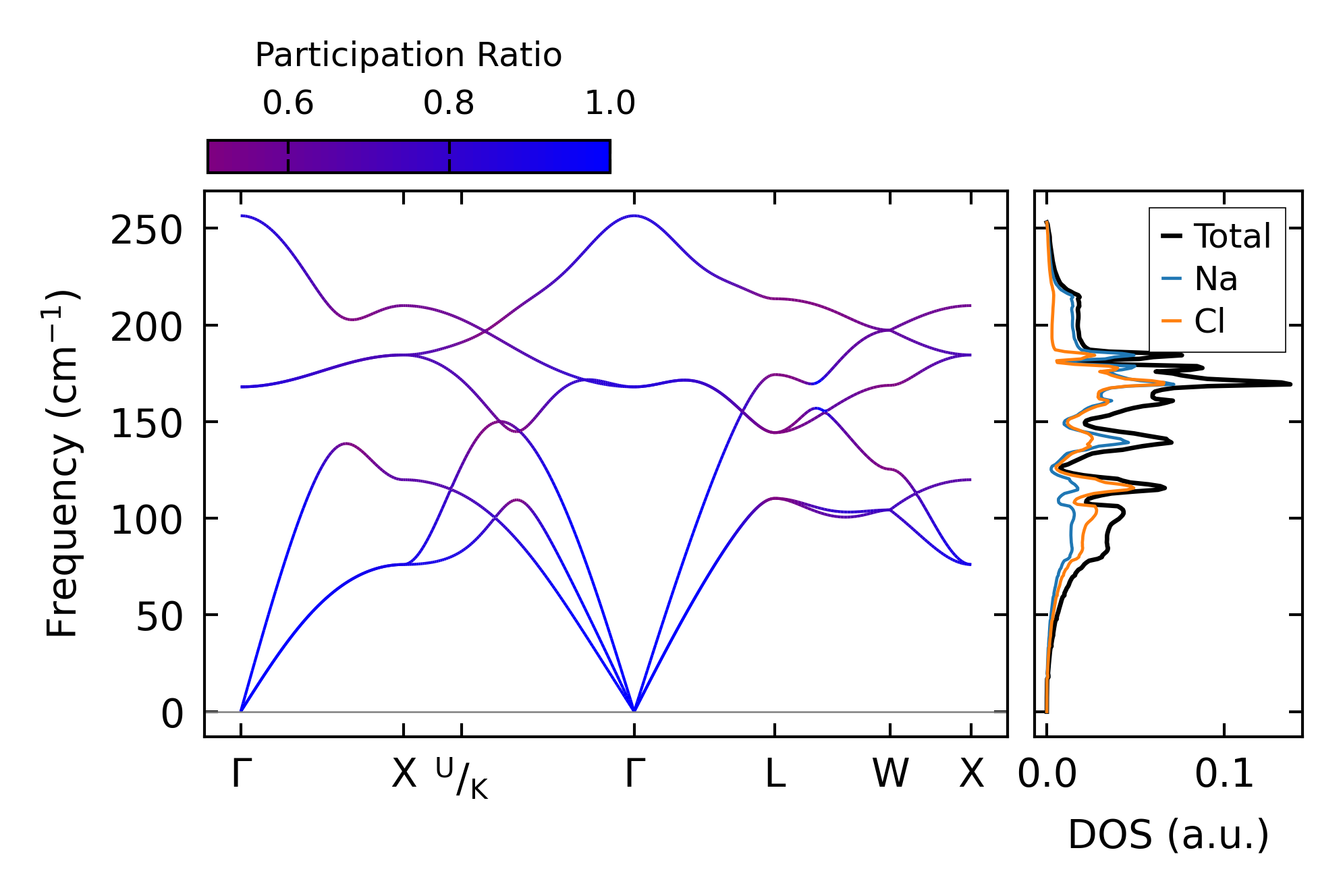
Fig. 5.2.1 Phonon dispersion with participation ratio and DOS of Silicon
Plot force constants
#!/usr/bin/env python3
# Filename: plot_fcs.py
import os
import argparse
import glob
from auto_kappa.io import FCSxml
from auto_kappa.plot.initialize import set_matplot, set_axis
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
def main(options):
fontsize = 7
fig_width = 2.8
aspect = 1.0
dpi = 600
## Prepare figure frame
set_matplot(fontsize=fontsize)
fig = plt.figure(figsize=(fig_width, aspect*fig_width))
plt.subplots_adjust(hspace=0.05)
axes = []
for ifig in range(2):
axes.append(plt.subplot(2, 1, ifig+1))
## Get file name
from plot_bandos import get_formula
formula = get_formula(options.base_dir)
line = f"{options.base_dir}/cube/*/{formula}.xml"
try:
file_fc3 = glob.glob(line)[0]
except IndexError:
print(" No FCS file found")
return
## Read FCS file
fcs = FCSxml(file_fc3)
## Plot FCs
fcs.plot_fc2(axes[0])
fcs.plot_fc3(axes[1])
axes[0].tick_params(labelbottom=False)
## Save figure
figname = "fig_fcs.png"
fig.savefig(figname, dpi=dpi, bbox_inches='tight')
print(" Output", figname)
print("FC2:", fcs.fc2.shape)
print("FC3:", fcs.fc3.shape)
if __name__ == '__main__':
parser = argparse.ArgumentParser(description='Input parameters')
parser.add_argument('--base_dir', dest='base_dir', type=str,
default="./mp-22862", help="directory of auto-kappa output")
args = parser.parse_args()
main(args)
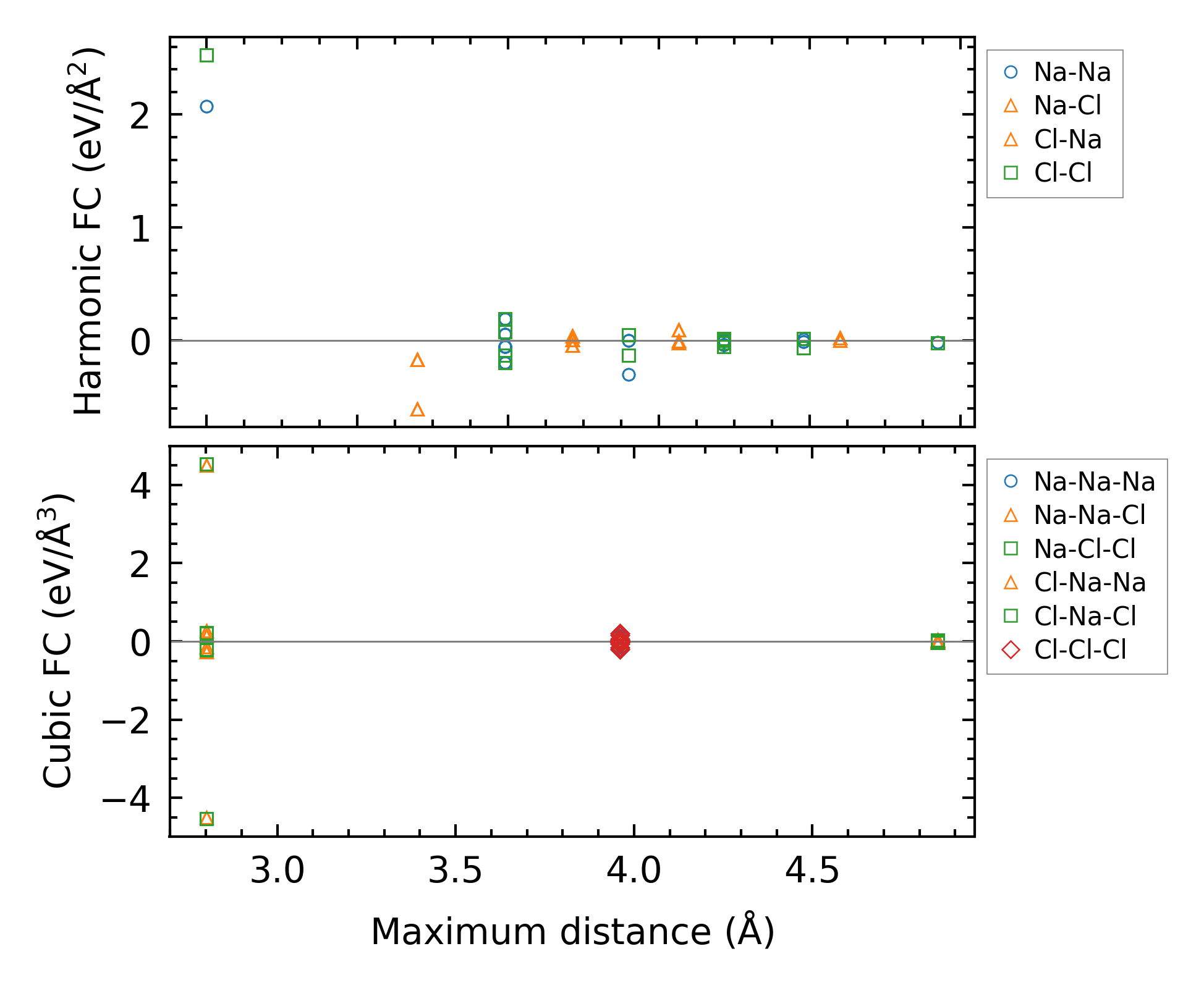
Fig. 5.2.2 Force constants of NaCl